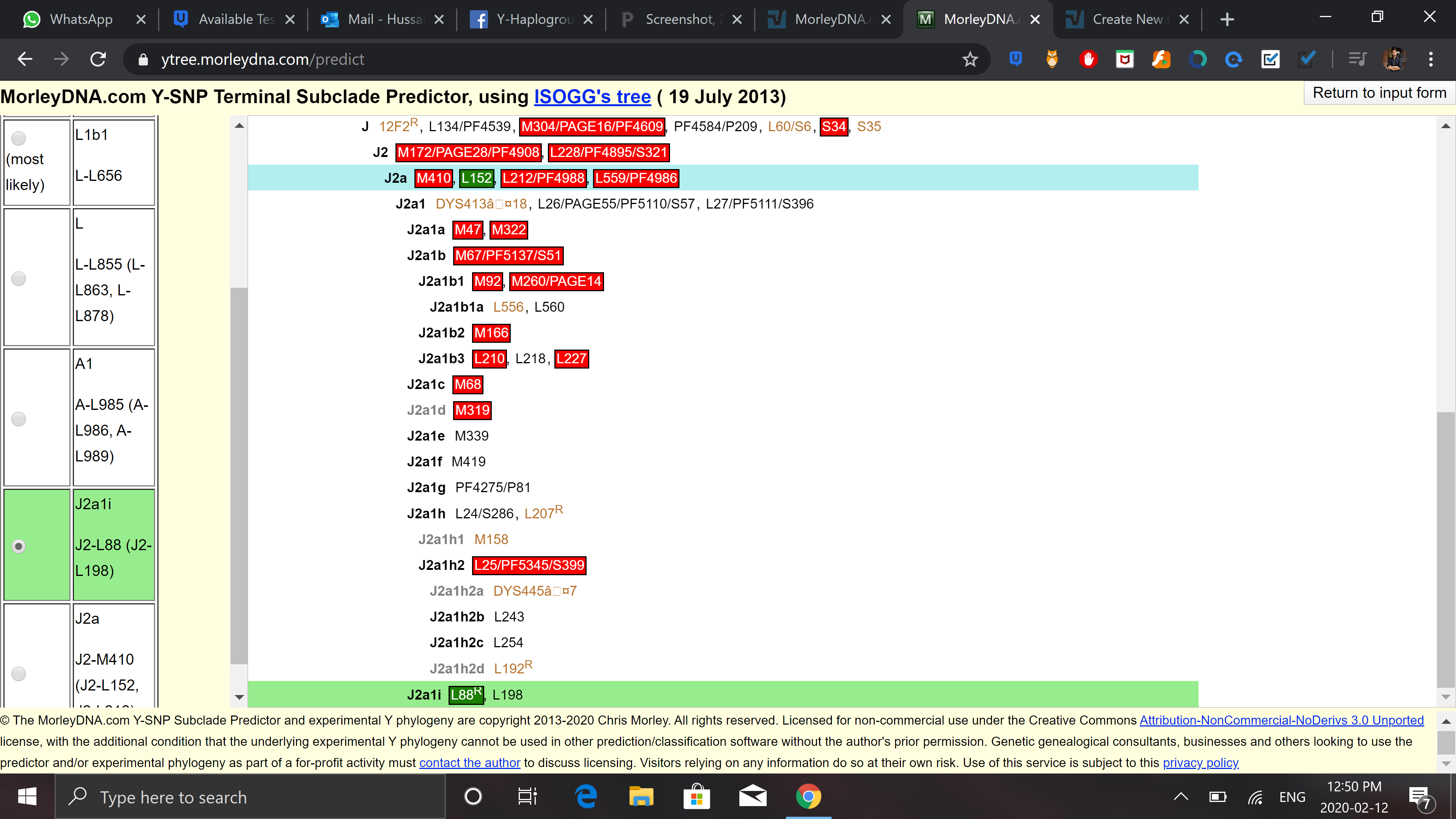
Alright so here is prediction from morley:
https://paste.pics/3ec3c7f1fc8322ccfcc01917894bbe2a
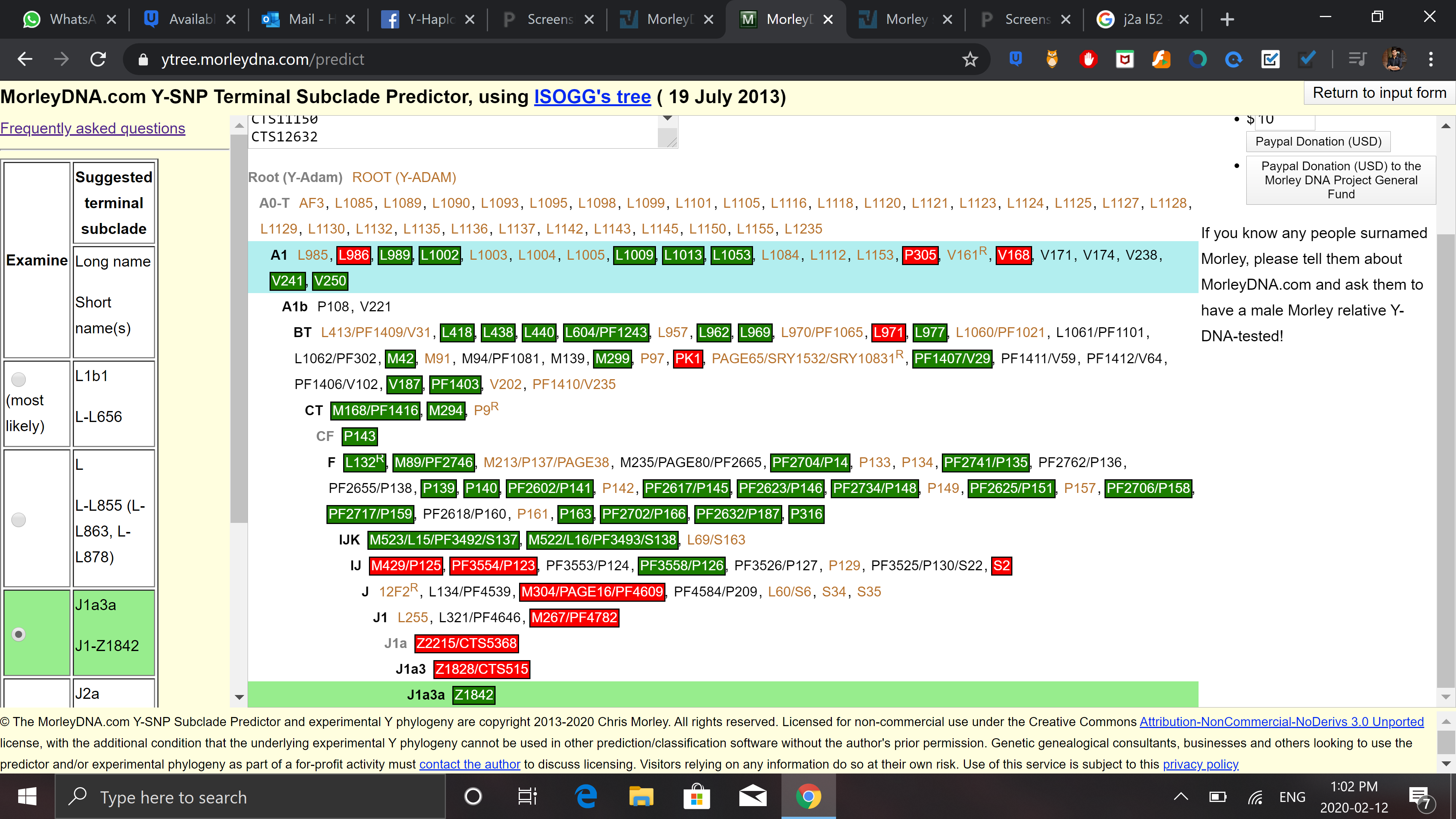
https://paste.pics/8081K
And by this my most likely is L1b1 even though i tested negatively for M317, M349 and M274. This should mean that the result by morly is wrong. Also, i tested negatively for L1a(M27, M76, P329 ) and L2 (L595) which only leaves me with L1c (M357) for which i haven't been tested positive or negative, which is common in Pashtuns and Baloch (some say jatt population as well so if someone knows anything on Jatts and which subclade of L they have, that would be great too).
So my, question is, if we look at the data above, it is logical to conclude that I can only be L1c since i am not tested either positive or negative for it?
Also, is it possible that I do not even belong to L? My next most probable haplogroups are as follows:
Any help would be appreciated.
https://paste.pics/3ec3c7f1fc8322ccfcc01917894bbe2a
https://paste.pics/8081K
And by this my most likely is L1b1 even though i tested negatively for M317, M349 and M274. This should mean that the result by morly is wrong. Also, i tested negatively for L1a(M27, M76, P329 ) and L2 (L595) which only leaves me with L1c (M357) for which i haven't been tested positive or negative, which is common in Pashtuns and Baloch (some say jatt population as well so if someone knows anything on Jatts and which subclade of L they have, that would be great too).
So my, question is, if we look at the data above, it is logical to conclude that I can only be L1c since i am not tested either positive or negative for it?
Also, is it possible that I do not even belong to L? My next most probable haplogroups are as follows:
Any help would be appreciated.
Comment